Study Results
The study has created a prototype of a sparse training tool that can train several LLMs of the Bert family The results show that the use of VENOM and Spatha can provide a sparse alternative to the traditional dense training procedure. The use of a Gradual Magnitude Pruning (GMP) strategy can generate the required semi-structured N:M sparsity pattern preserving the accuracy of a non-structured pruning technique. The sparse trained model, trained following a GMP strategy, achieves an F1 score of 87.40 after 50 epochs. This metric provides a measure of precision, 100 being the perfect score. Using equivalent standard magnitude pruning, we achieve an F1 score of 87.5. For context: magnitude pruning serves as our baseline method since it doesn’t place any restrictions on which weights in the model can be removed.
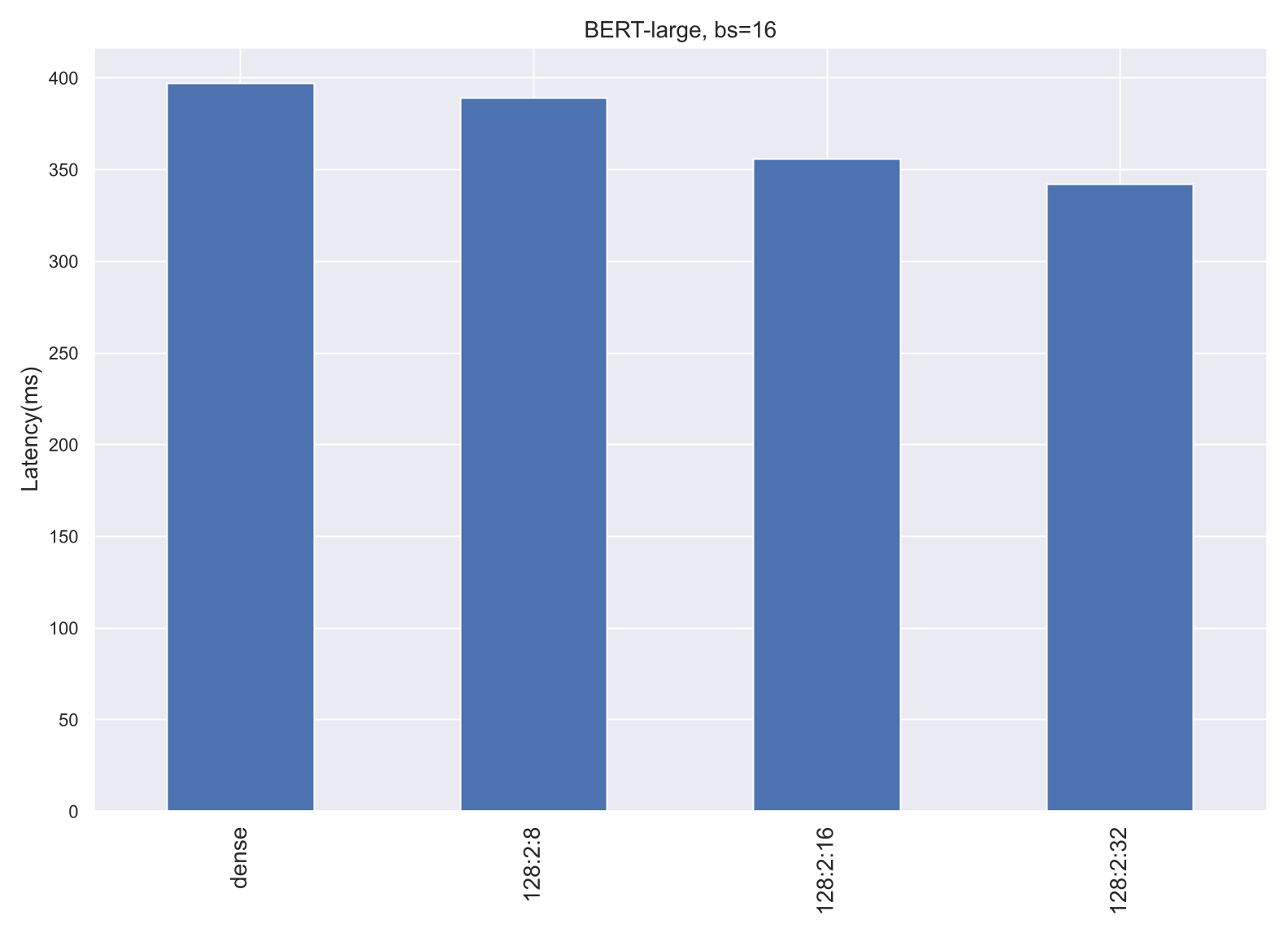
At the kernel level, Spatha kernels are up to 12x faster for the SDDMM kernel than their dense counterparts, and up to 37x faster for the SPMM kernel. Nevertheless, this translates into a modest 14% improvement of the Pytorch training step time (Figure [4]). The study has identified the inefficiencies that result in reduced translation of kernel speedup into training step speedups: (1) the current implementation requires additional data transformations to fit the ordering of the inputs, (2) the kernels are not tuned for all kinds of Nvidia GPUs, and (3) the data-exchange in distributed environments could be made in compressed storage format for sparse data structure for further efficiency.
This indicates a clear path to take the prototype to a production environment and serves as a showcase of the potential of sparse training.
The foundation of the sparse training tool is the VENOM compressed format of storage for sparse data structures and the Spatha implementation of the SPMM kernel linked to VENOM (https://github.com/UDC-GAC/venom ). Both developments exploit sparse models following an N:M generic sparsity pattern, accelerating the numerical kernels by taking advantage of Nvidia Sparse Tensor Cores, available on Nvidia GPUs since the Ampere architecture. The kernels are implemented in CUDA, and the SparseML library (https://github.com/neuralmagic/sparseml) was used to facilitate the access to sparsification recipes and the management of the associated masks and sparsified data structures. The code generated is publicly available in this repository: https://github.com/diegoandradecanosa/venom-training
The SparseML-based tool being the final form of our sparse training prototype.
As evident from the abovementioned repository, we built a support tool called Amdhal4ML to analyze the weight of the different numerical kernels in the training step of a model: amdhal4ML folder in the reposoitory. The Spatha kernels used in this work are located in the sparse_kernels folder of the repository. We also generated a set of performance analysis studies whose support code is contained in the performance_analysis folder. Before we had the final prototype, we used several scripts to perform tests to check the numerical correction of the kernels, and the feasibility and numerical correction of the integration of the kernels in a training step. This is located in the test_and_benchmarks folder. Finally, the main prototype is provided integrated in the SparseML set of libraries (sparseml folder). The repository contains several explanatory READMEs.
Benefits
While the study addressed specifically the needs of LLM training, the selected approach has a much broader potential impact as the GEMM (General Matrix-Matrix Multiplication) operation forms the core of most machine learning models, not just transformers. In that sense, the main challenge nowadays concerns LLMs, but the solution developed has the potential to be applied to future generations of ML models. Our study highlights the potential to accelerate and optimize the training of LLMs by leveraging the GPU hardware that underpins extreme-scale computing architectures. This acceleration is achieved using Nvidia’s Sparse Tensor Cores, specialized components designed to efficiently handle matrix operations with semi-structured N:M sparsity patterns. By exploiting this form of sparsity, we can significantly reduce the computational workload, leading to substantial improvements in training speed, and ultimately, reductions in both energy consumption and operational cost
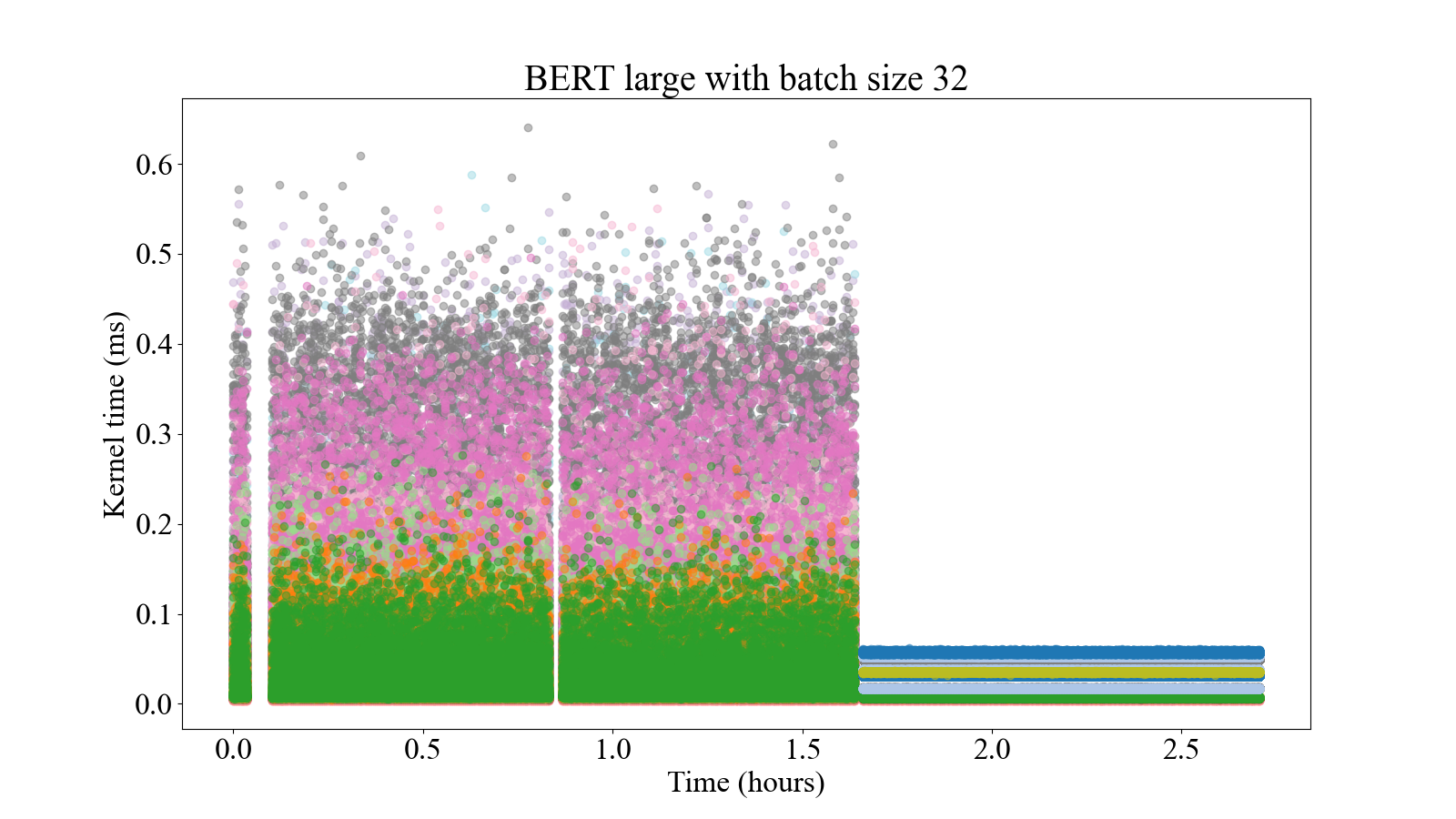
The study developments facilitate different trade-offs between more target model accuracy (less sparsity) and less training time (more sparsity) by configuring different sparsity levels. The sparse versions of the kernels are also more stable, presenting a less variable execution time (Figure [5]). In distributed environments, sparse training can take advantage of the use of an increasing number of GPUs, presenting similar speedups to dense trainings. Also, the final model will require less computational resources in inference time when it is pruned more aggressively, which can be seen as a second-order benefit of the sparse training.
Partners
| University of A Coruña (HPC expert): The Computer Architecture Research group (https://gac.udc.es/index.php/en/frontpage-english/) from University of A Coruña is a research group focused mainly in the area of High-Performance Computing (HPC). The area of research goes from: large-scale distributed systems applications, programming of kernels for heterogeneous platforms, architecture-level optimizations and programmability improvement. |
| CESGA (HPC provider):The Fundación Pública Gallega Centro de Supercomputación de Galicia (CESGA) (https://www.cesga.es) is a supercomputing center integrated into the Spanish Supercomputing Network (RES) that is a National Research Facility. CESGA runs one of the most powerful supercomputers in Spain (Finisterrae III) that is composed of 354 nodes, with 22,656 cores Intel Xeon Lake 8352Y, 128 GPUs NVIDIA A100, 118TB of memory, and 5000 TB online storage. These nodes are connected using Infiniband HDR. The peak capacity is 4PFlops. |
Team
- Diego Andrade
- Roberto López Castro.
- Juan Touriño
- Basilio Fraguela
- Diego Teijeiro
- José Carlos Mouriño
- Jorge Fernández Fabeiro.
- Francisco Javier Yáñez Rodríguez
Contact
Name: Diego Andrade Canosa
Institution: Universidade da Coruña, Spain
Email Address: diego.andrade@udc.es