Study Results
A suite of multi-GPU oriented implementations of the CG solver, both its standard algorithm and a pipelined version, was created as open-source software. The suite encompasses three types of communication libraries and can be used with both Nvidia and AMD GPUs.
Performance models were developed for the computational kernels in the CG method to verify that the single-GPU performance of the CG solver agrees with the theoretical expectation. For the inter-GPU communication overhead, i.e., peer-to-peer data exchange and all_reduce operations, standard micro-benchmarks and simple performance models were used to obtain a quantitative understanding of these costs. Time measurements of the new CG implementations were collected on several European supercomputers, showing better performance than state-of-the-art GPU implementations as well as scalable performance on more than 1000 GPUs.
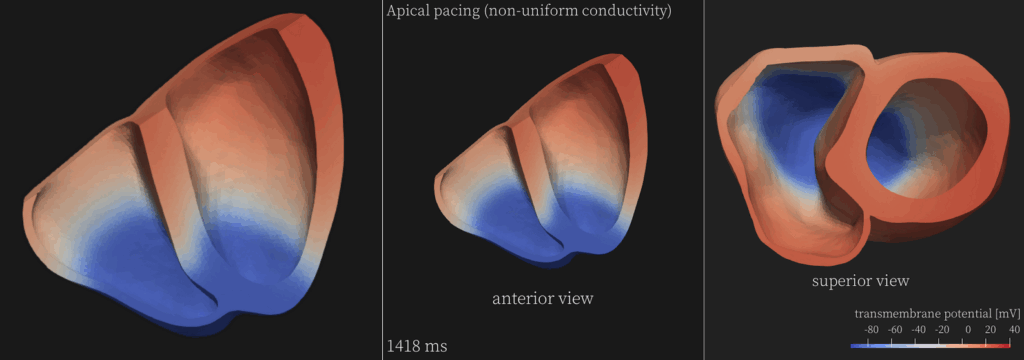
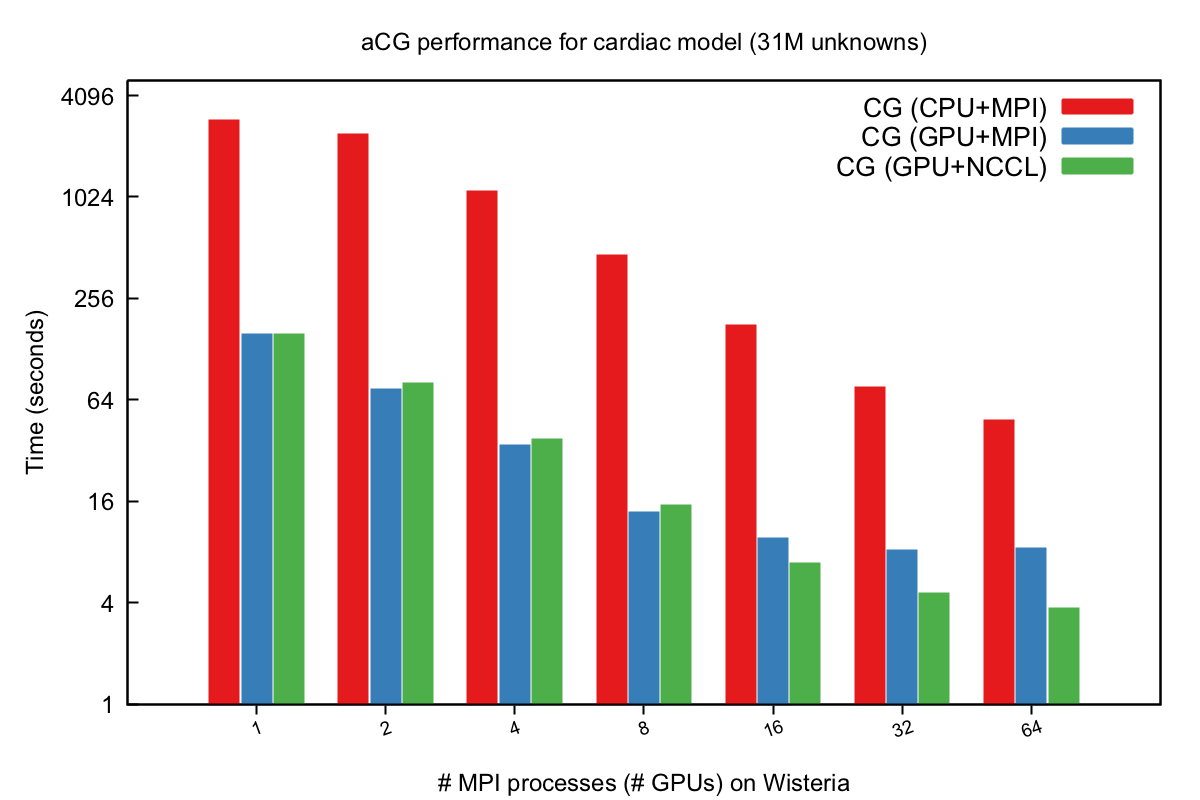
To test the parallel scalability of the multi-GPU implementations for large-scale, real-world scenarios, a simulator of cardiac electrophysiology and a simulator of porous-media flow were used with the original multi-CPU CG solvers replaced by the newly developed multi-GPU code. For example, using a sparse matrix arising from an unstructured tetrahedral mesh with 31 million unknowns for the cardiac simulator, the best multi-GPU performance using 64 Nvidia A100 GPUs was found to be 13 times faster than using 16 Intel Xeon Platinum 8360Y (36-core Ice Lake) CPUs. Moreover, the new multi-GPU implementations were found to achieve a speedup of 1.29x over the multi-GPU counterparts in the state-of-the-art linear-algebra library: PETSc.
Three types of communication libraries were adopted, namely, GPU-aware MPI, Nvidia’s NCCL or AMD’s RCCL, and OpenSHMEM-based one-sided communication libraries. These communication libraries were used to implement the standard CG algorithm and a pipelined version of CG for the purpose of efficiently using large clusters of Nvidia or AMD GPUs. Care was also taken to remove the unnecessary synchronization points and CPU-GPU data transfers.
Benefits
The suite of new CG implementations developed in this innovation study enables a systematic investigation of the three types of state-of-the-art communication libraries that target multi-GPU systems. Their respective advantages and disadvantages were studied quantitatively. In particular, one-sided communication that is provided by OpenSHMEM based libraries, and its application in implementing Krylov subspace methods (with CG as the best-known example), is a topic that has so far only scarcely researched by the HPC community. The findings of this study can thus provide useful recommendations to the maintainers/developers of modern linear-algebra software libraries.
The parallel efficiency of the new CG implementations was studied on several cutting-edge European supercomputers. We showed that the new implementations are faster than the GPU implementations in a state-of-the-art software library. Moreover, the new CG implementations using one-sided communication achieved scalable performance on more than 1000 GPUs. These results may encourage more adoption of GPU computing in various branches of computational science.
Partners
| Koç University (Turkey) provided HPC expertise. The PI was Prof. Didem Unat, together with Sinan Ekmekçibaşı, Doğan Sağbili, and Emre Düzakın as participants in the study. |
| Simula Research Laboratory (Norway) provided both HPC expertise and domain knowledge of computational physiology, as well as a CPU implementation of a real-world demonstrator that simulates the electrophysiology of the heart. The PI was Prof. Xing Cai, together with Dr. James Trotter and Dr. Johannes Langguth as participants in the study. |
Contact
Name: Didem Unat, Xing Cai
Institution: Koç University, Simula Research Laboratory
Email Address: dunat@ku.edu.tr , xingca@simula.no